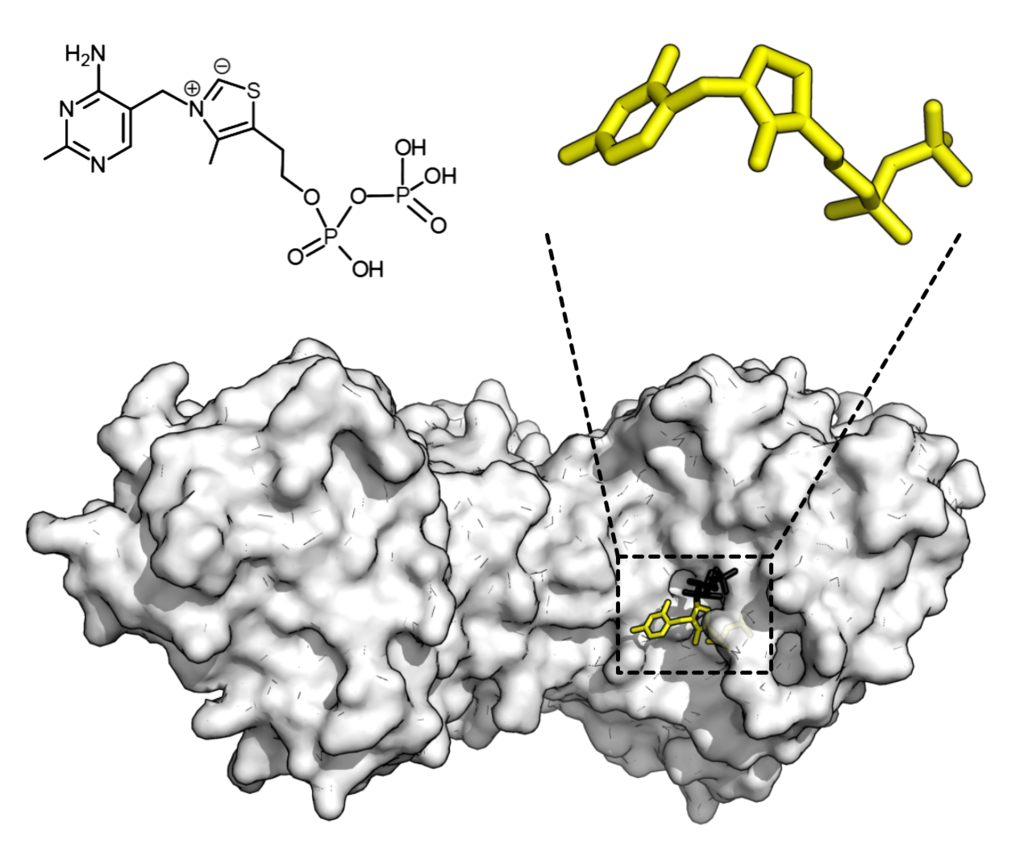
Chemical structure for thiamine pyrophosphate and protein structure of transketolase. Cofactor thiamine pyrophosphate in yellow and substrate xylulose 5-phosphate in black. Credit: Thomas Shafee/Wikipedia
Scientists have prototyped a new method for “rational engineering of enzymes” to deliver improved performance. They have created an algorithm, which takes into account the evolutionary history of an enzyme, to mark where mutations can be introduced with a high probability of providing functional improvements.
Their work – published today in the magazine Nature Communications— can have significant and far-reaching impacts on a variety of industries, from food production to human health.
Enzymes are essential to life and key to the development of medicines and innovative tools to address societal challenges. They have evolved over billions of years through changes in the amino acid sequence that supports their 3D structure. Like beads on a string, each enzyme consists of a sequence of several hundred amino acids that encode its 3D shape.
With one of 20 possible amino acid “beads” at each position, there is a huge variety of possible sequences in nature. By forming their 3D shape, enzymes perform a specific function such as digesting our dietary proteins, converting chemical energy into power in our muscles, and destroying bacteria or viruses that invade cells. If you change the sequence, you can disrupt the 3D shape and this usually changes the functionality of the enzyme, sometimes rendering it completely ineffective.
Finding ways to improve enzyme activity would be extremely useful for many industrial applications and, using modern tools in molecular biology, it is simple and cost-effective to engineer changes in amino acid sequences to facilitate improvements in enzyme performance. theirs. However, random introduction of three or four changes in sequence can lead to a dramatic loss of their activity.
Here, scientists report a promising new strategy to rationally engineer an enzyme called “beta-lactamase.” Instead of introducing random mutations into a distribution approach, researchers at the Broad Institute and Harvard Medical School developed an algorithm that takes into account the enzyme’s evolutionary history.
“At the heart of this new algorithm is an evaluation function that utilizes thousands of beta-lactamase sequences from many different organisms. Instead of a few random changes, up to 84 mutations in a sequence of 280 were designed to improve functional performance .” said Dr. Amir Khan, Associate Professor in the School of Biochemistry and Immunology at Trinity College Dublin, one of the co-authors of the research.
“And surprisingly, the newly designed enzymes had improved activity and stability at higher temperatures.”
Eve Napier, a second-year Ph.D. student at Trinity College Dublin, determined the experimental 3D structure of a newly designed beta-lactamase, using a method called X-ray crystallography.
Its 3D map revealed that despite changes in 30% of the amino acids, the enzyme had an identical structure to wild-type beta-lactamase. He also discovered how coordinated changes in amino acids, introduced simultaneously, can efficiently stabilize the 3D structure – in contrast to the individual changes that usually damage the structure of the enzyme.
Eve Napier said, “Overall, these studies reveal that proteins can be engineered for enhanced activity by dramatic ‘jumps’ in new sequence space.
“The work has a wide range of applications in industry, in processes that require enzymes for food production, enzymes for the degradation of plastics and those related to human health and disease, so we are quite excited about the future possibilities.”
More information:
Nature Communications (2024). DOI: 10.1038/s41467-024-49119-x
Provided by Trinity College Dublin
citation: Scientists invent algorithm to engineer improved enzymes (2024, June 20) retrieved June 20, 2024 from https://phys.org/news/2024-06-scientists-algorithm-enzymes.html
This document is subject to copyright. Except for any fair agreement for study or private research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.